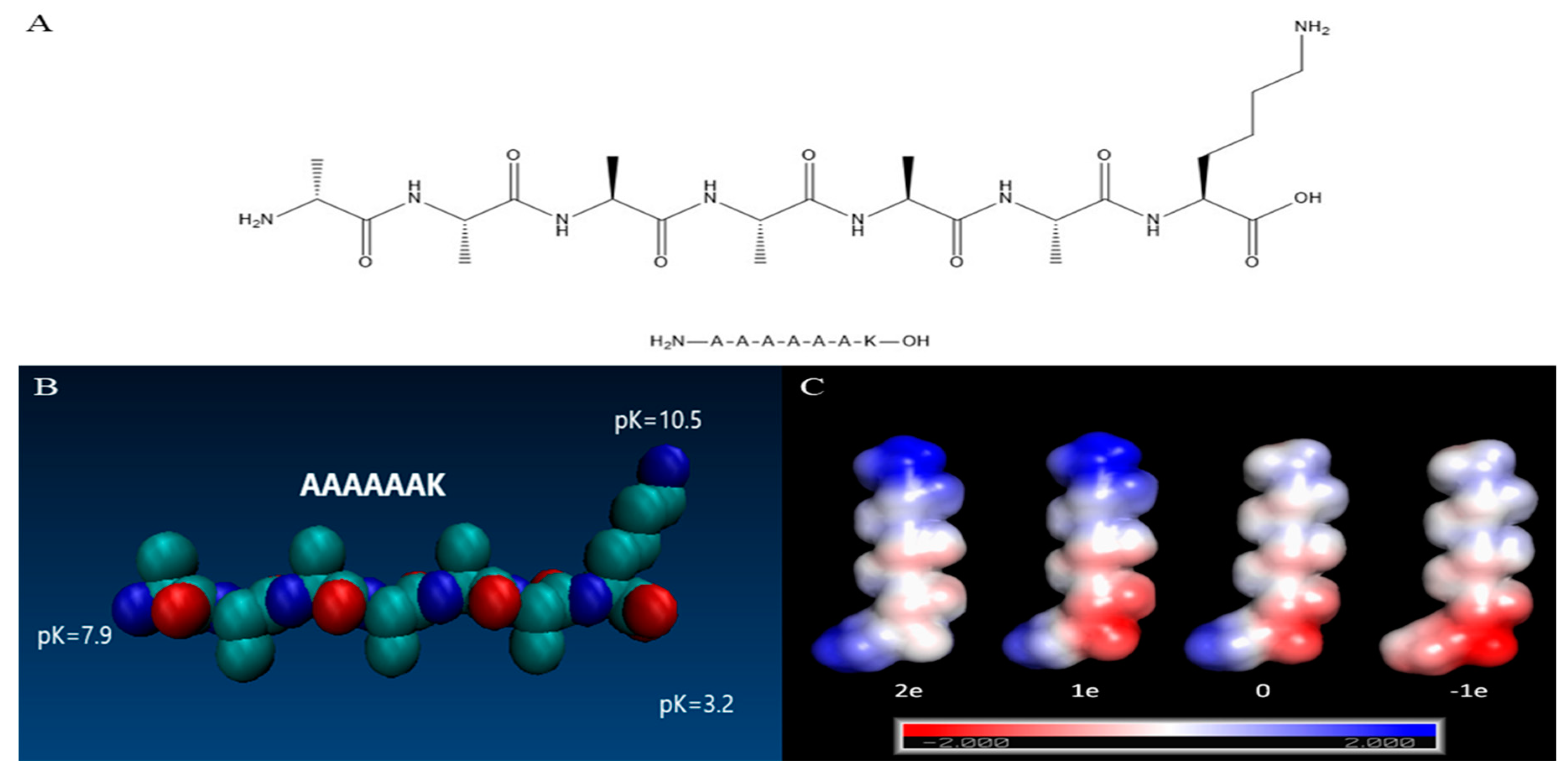
Molecules | Free Full-Text | Self-Assembling Behavior of pH-Responsive Peptide A6K without End-Capping

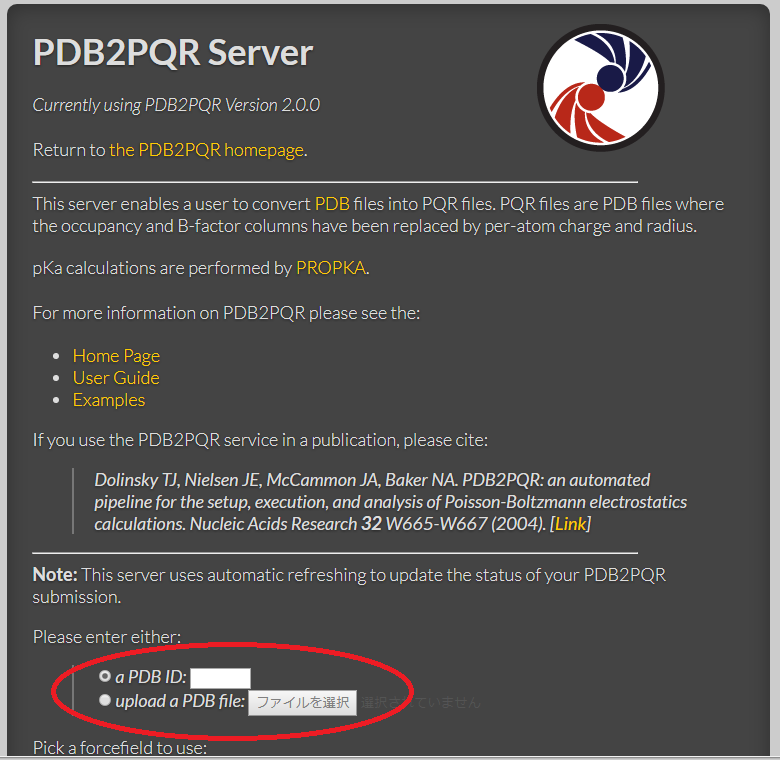
PDF) Web servers and services for electrostatics calculations with APBS and PDB2PQR | Wilfred Li - Academia.edu
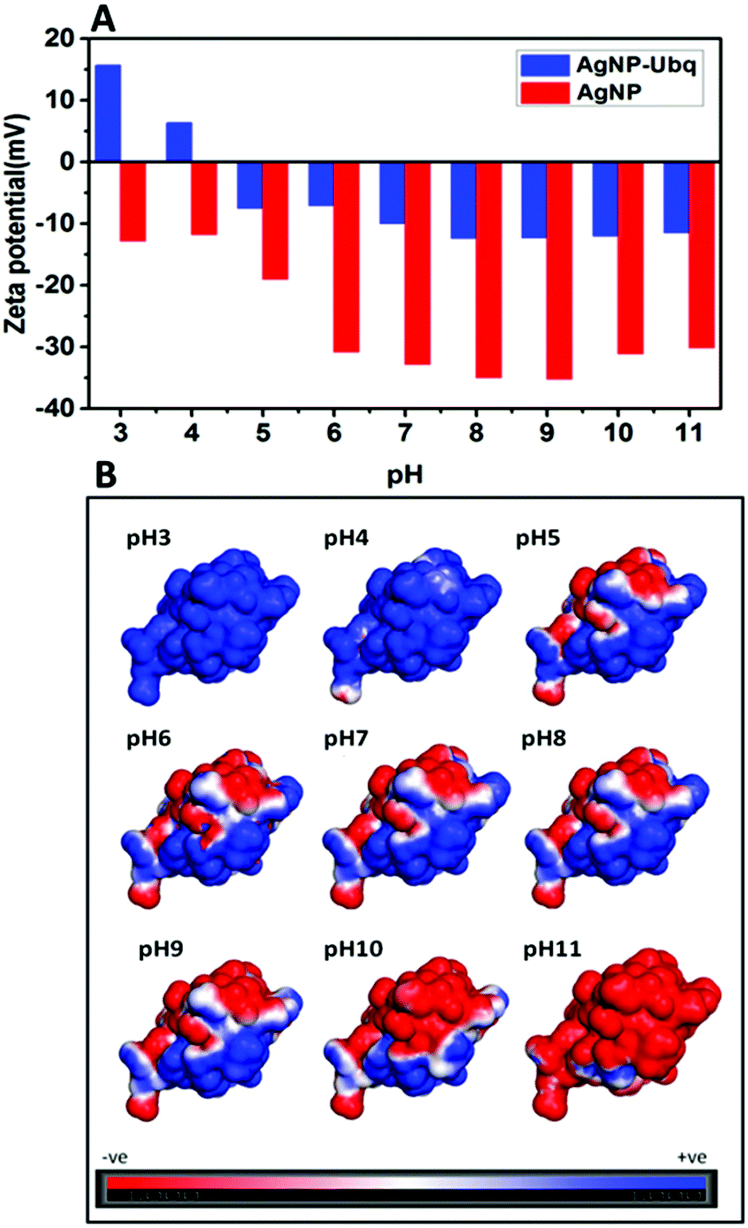
An ultrastable conjugate of silver nanoparticles and protein formed through weak interactions - Nanoscale (RSC Publishing) DOI:10.1039/C5NR03047A
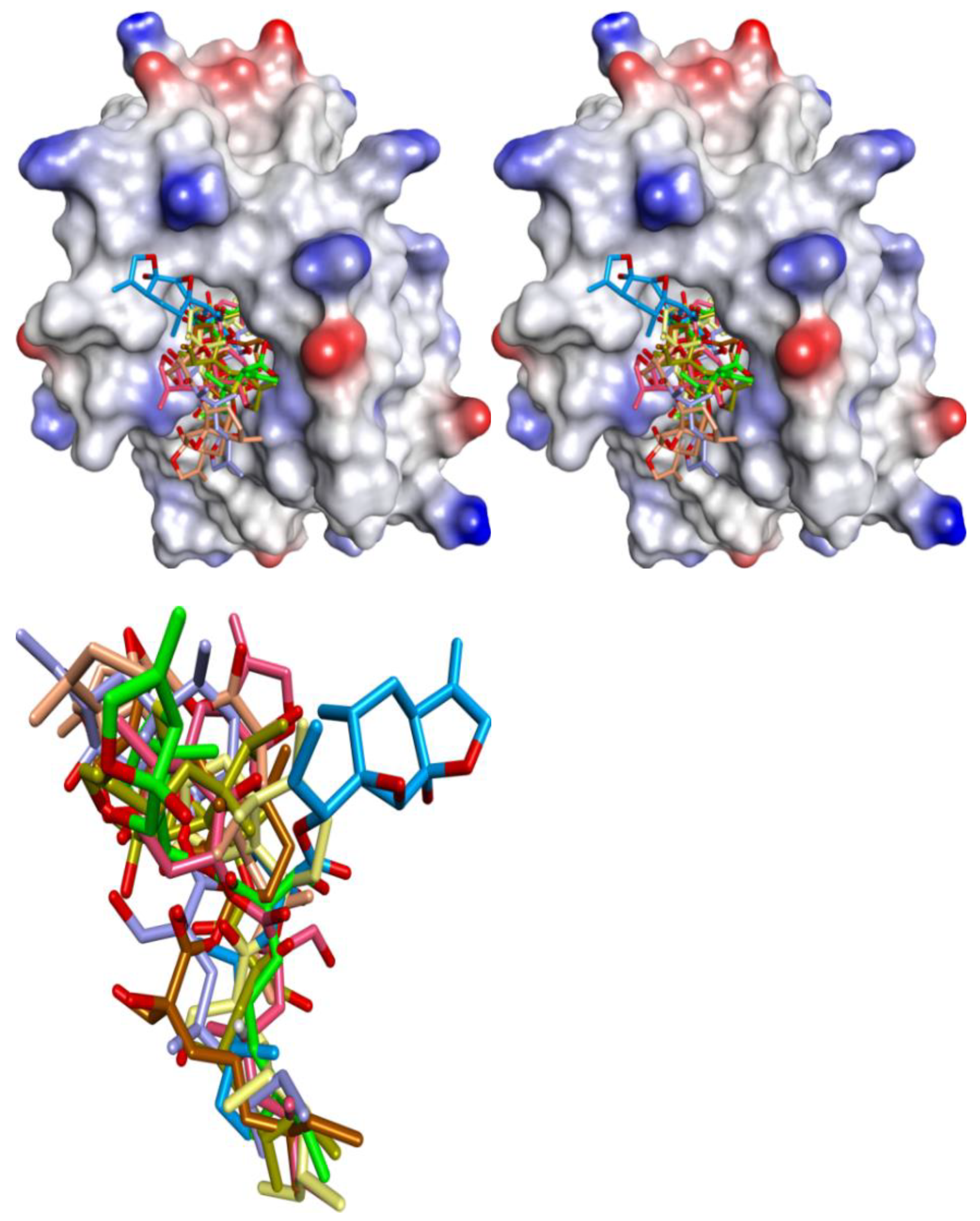
Chemistry | Free Full-Text | Natural Products as Mcl-1 Inhibitors: A Comparative Study of Experimental and Computational Modelling Data

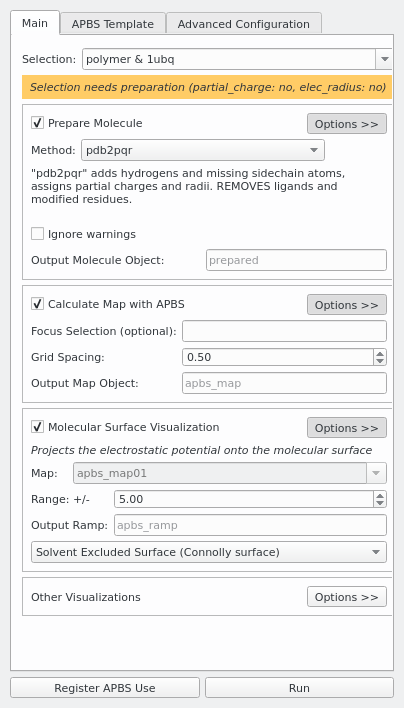
Improvements to the APBS biomolecular solvation software suite - Jurrus - 2018 - Protein Science - Wiley Online Library

AMOEBA Force Field Trajectories Improve Predictions of Accurate pKa Values of the GFP Fluorophore: The Importance of Polarizability and Water Interactions | The Journal of Physical Chemistry B

Improvements to the APBS biomolecular solvation software suite - Jurrus - 2018 - Protein Science - Wiley Online Library

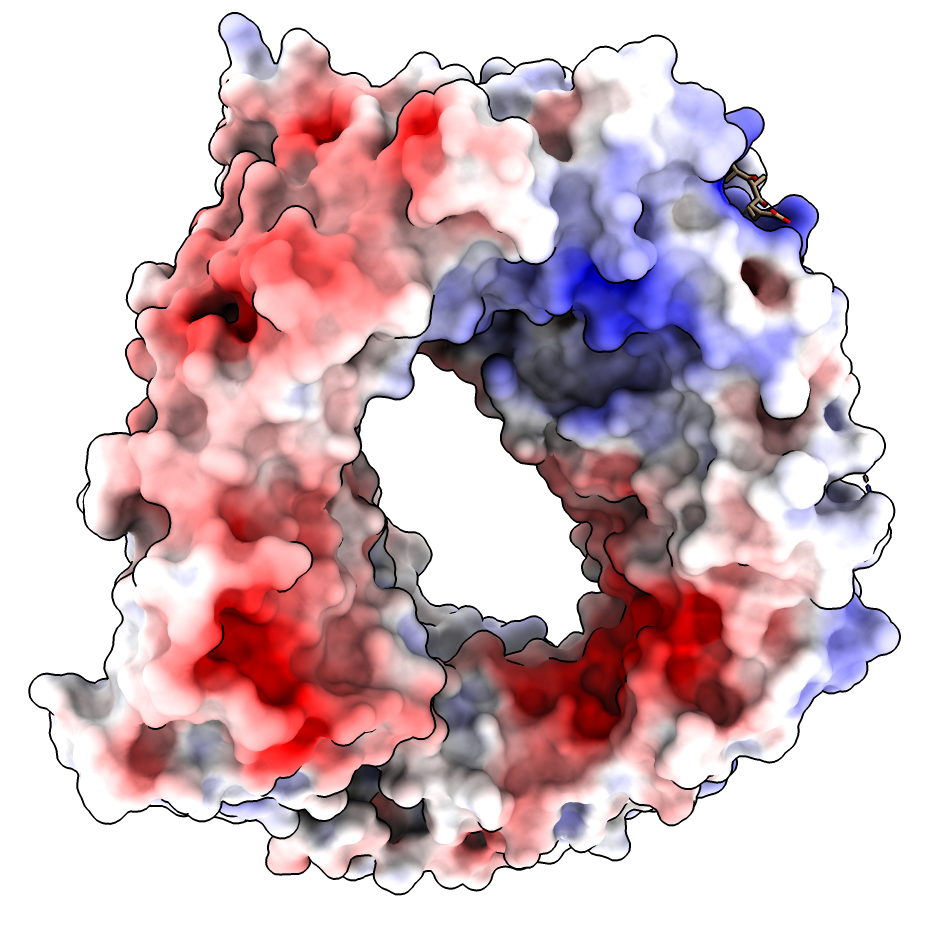
Electrostatic potential map of the Cqm1 dimer. ± 15 kT/e electrostatic... | Download Scientific Diagram
GitHub - Electrostatics/electrostatics.github.io: APBS & PDB2PQR - software for biomolecular electrostatics and solvation

Improvements to the APBS biomolecular solvation software suite - Jurrus - 2018 - Protein Science - Wiley Online Library

Improvements to the APBS biomolecular solvation software suite - Jurrus - 2018 - Protein Science - Wiley Online Library
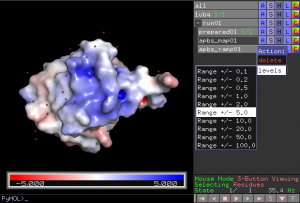
![PyMOL] side-by-side comparison of 3 electrostatic surface potentials | My software notes PyMOL] side-by-side comparison of 3 electrostatic surface potentials | My software notes](https://kpwu.files.wordpress.com/2012/09/1z66.png)